CRISPR/Cas9
Carbomenu has protein expression platform in both prokaryotic cells (E.coil) and eukaryotic cells (Yeast/CHO-k1/293). Carbomenu is able to deal with kilogram level customized protein production orders. Our product line can be optimized according to customer orders to meet personalized needs.
Information
Known as: | CRISPR associated protein 9 |
Species: | S. pyogenes |
Cat.No.: | CM4005 |
Nuclear localization signal: | With NLS or without NLS |
Expression Host: | Ecoli. |
Package: | 500pmol、1000pmol、5000pmol |
Price: | |
500pmol | Inquiry |
1000pmol | Inquiry |
5000pmol | Inquiry |
Introduction
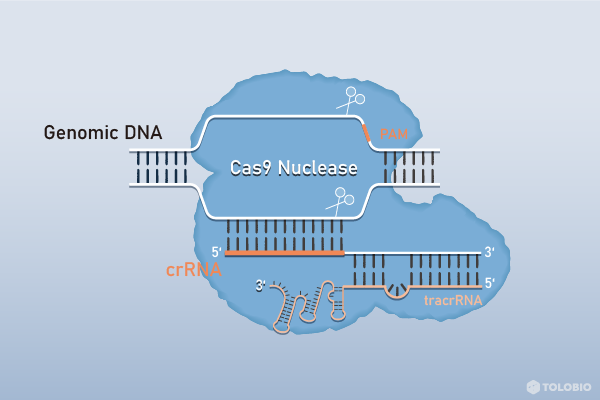
Cas9 nuclease is an RNA-mediated endonuclease from S. Pyogenes. Cas9 cleaves specifically double-stranded DNA (or, in the presence of DNA PAM, single-stranded DNA or single-stranded RNA). Cas9 cleavage site is located in the target sequence, 3 bases away from PAM (NGG) region. PAM is necessary for Cas9 identification and cutting. Because the reaction requires the addition of RNA (sgRNA or crRNA:tracrRNA), the entire system including the Cas9 enzyme, must be free of any nuclease activity.
HNH and RuvC domains in Cas9 are responsible for slicing sgRNA paired and unpaired strands in target DNA, respectively. Both H840 and D10 of Cas9 were mutated to A, which inactivated both HNH and RuvC domains of Cas9 to get dCas9 protein. Therefore, dCas9 has only binding target DNA activity, but no cleavage target DNA activity.
Product Component
Package | PKG1 | PKG2 |
Cas9 Nuclease | 500pmol | 1000pmol |
10×Cas9 Buffer | 1mL | 1mL×2 |
Control Target DNA and sgRNA | 2 rxn | 2 rxn×2 |
Recommended reaction system(20ul)
Component | Volume |
10×Cas9 Buffer | 2μL |
250 nM sgRNA | 2μL |
1μM Cas9 Nuclease | 1μL |
30 nM substrate DNA | 2μL |
Nuclease-free water up to | 20μL |
Incubate at 37 °C for 1 h, 85 °C 10 min | |
FOR RESEARCH USE ONLY
References
1.Genome engineering using the CRISPR-cas9 system. F Ann Ran, Patrick D Hsu, Jason Wright, Vineeta Agarwala,et al. Nature Protocols volume 8, 2281–2308 (2013).
2.CRISPR/Cas9-mediated viral interference in plants. Ali, Z., Abulfaraj, A., Idris, A., Ali, S., Tashkandi, M., and Mahfouz, M. M. (2015). Genome Biol. 16, 238–249. doi: 10.1186/s13059-015-0799-6.
3.Gene Editing and Crop Improvement Using CRISPR-Cas9 System.Leena Arora and Alka Narula. Front. Plant Sci., 08 November 2017. doi.org/10.3389/fpls.2017.01932.
4.In vivo blunt-end cloning through CRISPR/Cas9-facilitated non-homologous end-joining. Geisinger, J. M., Turan, S., Hernandez, S., Spector, L. P., and Calos, M. P. (2016). Nucleic Acids Res. 44, e76. doi: 10.1093/nar/gkv1542.